User Guide¶
You have the choice between two classifiers: an SVM (default) or a random forest. By default the classifier will use predefined parameters but you can also use the gridsearch option to find new parameters. The analysis results will be saved by default in a results folder but you can also use the dedicated parameter to customize it.
Code installed from Github¶
Here are two examples of commands you can use if you installed the code from Github:
# Train an SVM with predefined parameters and save the analysis
# in the "analysis_results" folder
(activities) $ python run.py -model svm -gridsearch n -output-folder analysis_results
# Train a random forest with gridsearch and save the analysis in
# the "results" folder
(activities) $ python run.py -model rf -gridsearch y -output-folder results
Package installed from PyPI¶
Here are two examples of commands you can use if you installed the package from PyPI:
# Train an SVM with predefined parameters and save the analysis
# in the "analysis_results" folder
$ rrgp -model svm -gridsearch n -output-folder analysis_results
# Train a random forest with gridsearch and save the analysis in
# the "results" folder
$ rrgp -model rf -gridsearch y -output-folder results
For your reference, our tables and figures are repeated below, so you can check the reproducibility of our solution.
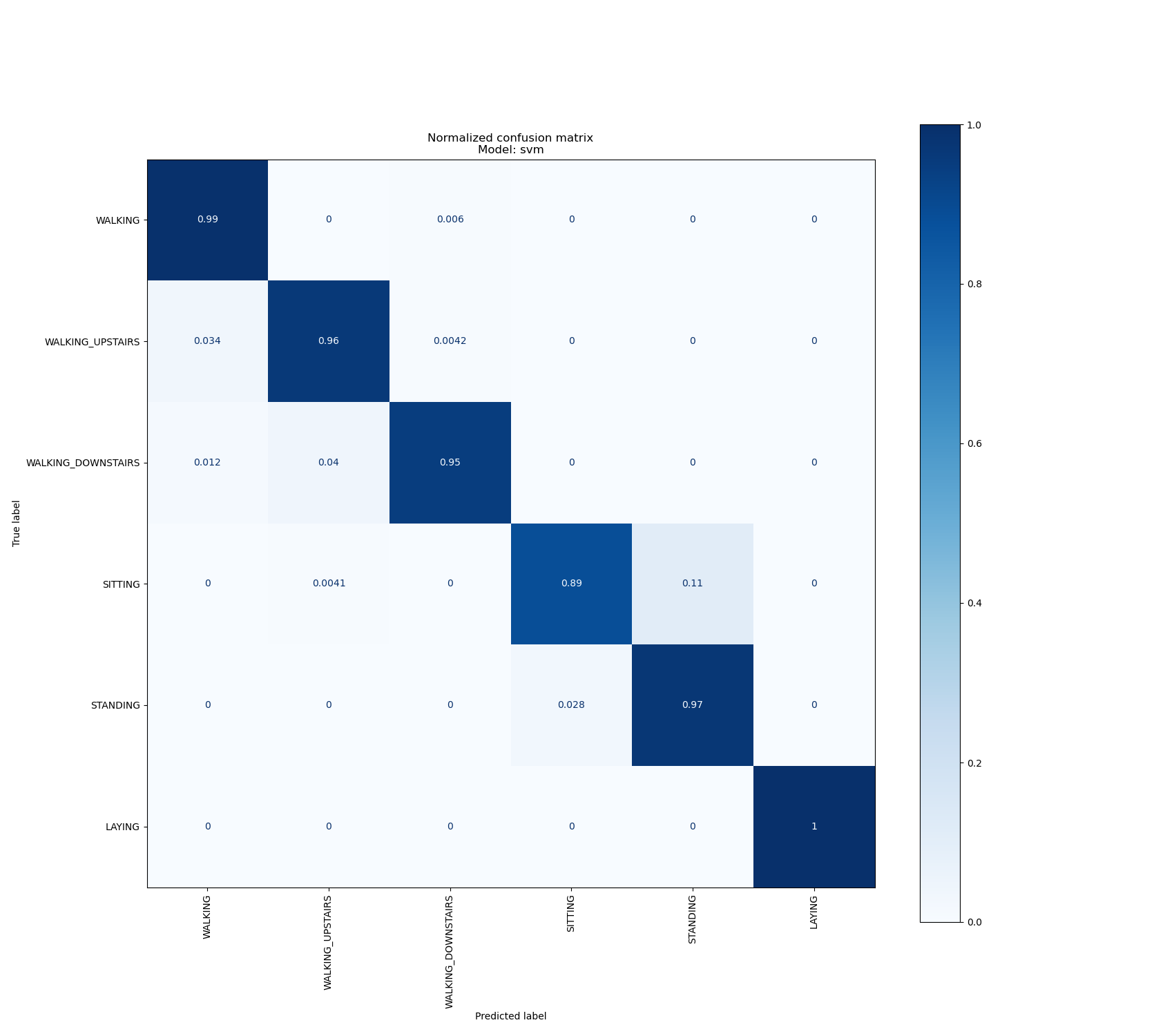
Results with the SVM model¶
For this experiment, we used an SVM model with the following parameters (kernel = “rbf”, gamma = 0.0001, C = 1000).
Precision (avg) |
Recall (avg) |
F1 score (avg) |
Accuracy |
|---|---|---|---|
0.961 |
0.961 |
0.961 |
0.961 |
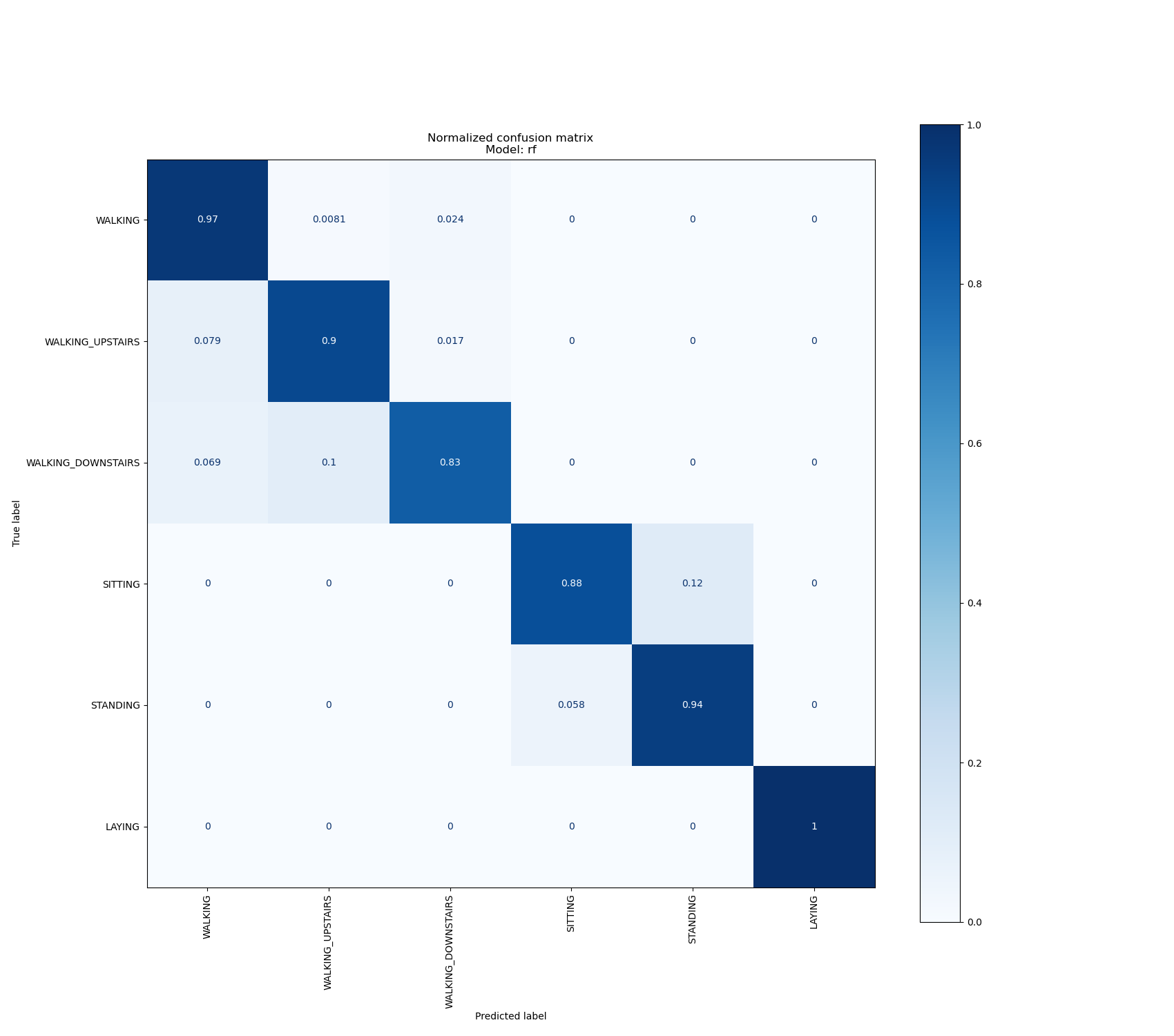
Results with the random forest model¶
For this experiment, we used a random forest model with the following parameters (n_estimators = 50, max_depth = 25, min_samples_split = 2, min_samples_leaf = 4, bootstrap = True).
Precision (avg) |
Recall (avg) |
F1 score (avg) |
Accuracy |
|---|---|---|---|
0.924 |
0.924 |
0.924 |
0.924 |
Customizing the parameters¶
If you want to customize the default parameters or the gridsearch ones, you can just modify them in the algorithm.py file.
Adding a new model¶
This package allows the use of two Scikit learn models: SVM and random forest. If you want to test another model, please follow this procedure:
Add a new option with the name of your model in the get_args function in run.py
Add an elif condition implementing your model in algorithm.py
You can then simply call the run.py script using your new model option.
Using custom Data¶
If you want, you can use custom data for either the training set, the test set, or both.
To do so, you have to specify the path of the data set .txt files (data and labels) in the arguments.
Here is an examples of command you can use to load a custom dataset:
# Train an SVM with predefined parameters and use custom train data
# and custom test data
(activities) $ python run.py -model svm -gridsearch n -train-data data/X_train.txt -train-labels data/y_train.txt -test-data data/X_test.txt -test-labels data/y_test.txt
Or if you installed the package from PyPI:
# Train an SVM with predefined parameters and use custom train data
# and custom test data
$ rrgp -model svm -gridsearch n -train-data data/X_train.txt -train-labels data/y_train.txt -test-data data/X_test.txt -test-labels data/y_test.txt
Dataset Formatting
Please have a look at the UCI HAR Dataset documentation for information about the formatting of the data. Here is a quick summary :
Data : 561 feature-columns (see
features.txtandfeatures_info.txt)Labels : from 1 to 6 : (see
activity_labels.txt)